Microbiology (JP)
この教室に関連するワード
- 口腔微生物
- 腸内細菌
- インフラマソーム
- 遺伝子発現調節
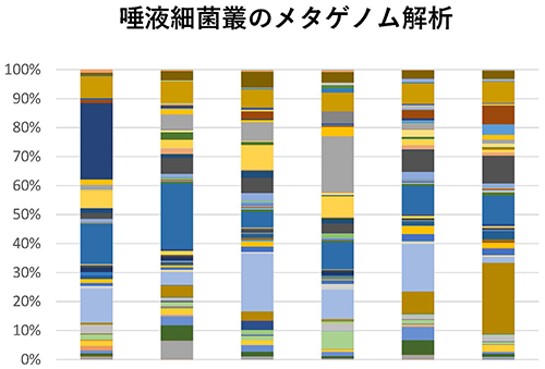
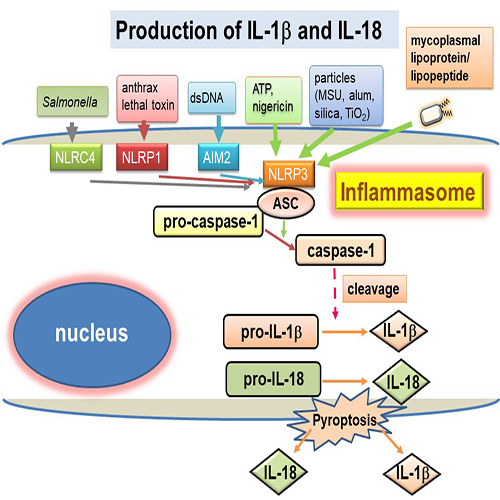
我々は、主に口腔微生物が腸内細菌叢に及ぼす影響、インフラマソームと炎症性疾患の関連性、および微生物ゲノム断片による真核細胞の遺伝子発現調節などを研究している。
ヒトの体には多数の細菌が存在し、宿主であるヒトの細胞と相互作用している。人間の細胞は一人あたりおよそ30兆個、また、腸内細菌だけで一人あたりおよそ38兆個存在すると考えられており、現在ではこれらが相互作用してひとつの生き物であるという概念も提唱されている。腸内細菌はこれまで考えられてきた以上に全身の状態に関わっていることがわかってきているが、我々は、口腔微生物を嚥下により常時取り込んでいることから、それらが腸内細菌に影響を与えるのではないかと考え、口腔微生物、特にCandida albicansが腸内細菌叢に与える影響について研究している。
インフラマソームは、NLR、ASCならびにprocaspase-1からなる三分子複合体で、多様な生理活性をもつ炎症性サイトカインのひとつであるIL-1βならびにIL-18 の産生を制御する細胞内センサーである。これまでに口腔細菌によるインフラマソーム活性化メカニズムについて明らかにしてきてた。さらにToll様受容体2のリガンドであるMycoplasma salivarium由来合成リポペプチドFSL-1を用いてインフラマソーム活性化の詳細なメカニズムを解析している。
微生物ゲノム断片による真核細胞の遺伝子発現調節については、我々はmRNAの5’ 非翻訳領域にmRNAの二次構造を変化させるような配列を導入することによりタンパク質の発現をシームレスに調節できるのではないかと考えている。実際に、微生物ゲノムDNA断片ライブラリーを5’ 非翻訳領域に挿入することにより、タンパク質発現量を0.5%から少なくとも300%程度までシームレスに調節できることが示唆されており、これについてに研究も行っている。
担当教員
Akira Hasebe

